About TBEP
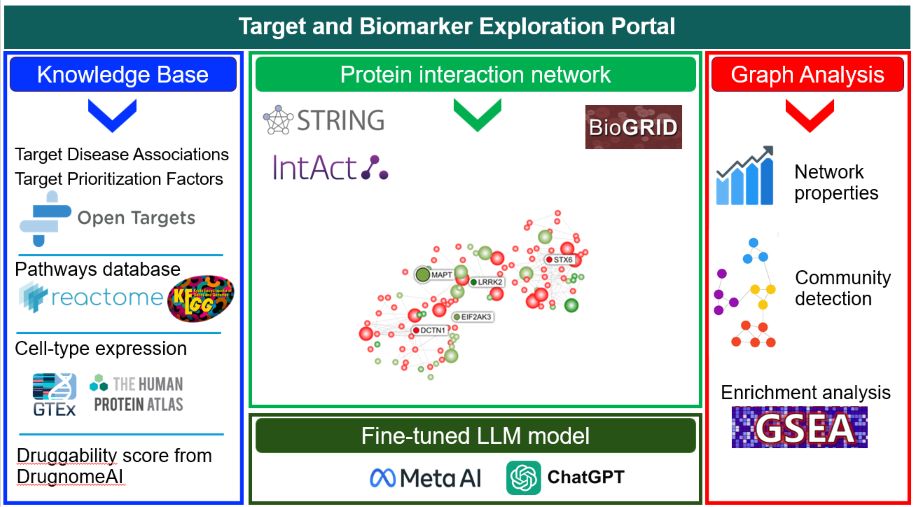
The Target and Biomarker Exploration Portal (TBEP) is a cloud-enabled, web-based bioinformatics platform designed to accelerate the identification of disease-associated targets and biomarkers. While conventional approaches often overlook key regulatory proteins, TBEP leverages network-based computational methods to model complex biological systems. By integrating multi-modal biomedical datasets—including human genetics, functional genomics, and curated protein-protein interaction (PPI) networks—TBEP provides a scalable, AI-driven framework for real-time analysis and visualization. This empowers researchers to uncover critical regulatory hubs and novel targets that traditional analyses may miss, supporting a broad range of applications.
Platform Architecture
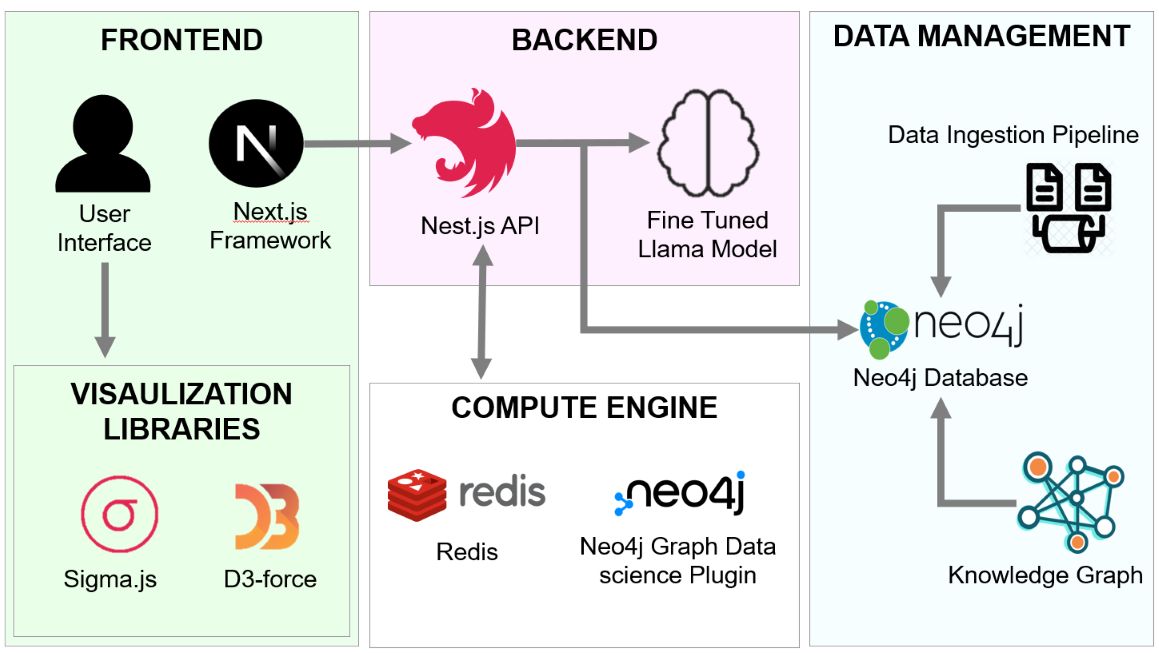
TBEP’s architecture is designed for robust, scalable, and interactive data exploration. It consists of three interrelated components: a dynamic frontend for visualization and user interaction, a powerful backend for data management and analysis, and a specialized Question-Answering (QA) system known as the TBEP Assistant. This integrated design allows for seamless data analysis and hypothesis generation.
Frontend and Visualization
The frontend is built with Next.js, ensuring a responsive and high-performance user experience. Network visualizations are powered by Sigma.js and D3-force, which use WebGL and physics-based layouts to render large, complex networks efficiently. This allows for real-time, interactive exploration of biological data, enabling users to dynamically adjust visualizations and identify significant patterns.
Backend and Data Management
The backend utilizes Nest.js with GraphQL for scalable application logic. Bioinformatics data is stored in Neo4j, a graph database that effectively models the interconnected nature of biological entities. This ensures rapid, flexible querying of large datasets sourced from curated databases like STRING, Reactome, and Open Targets. Graph algorithms are powered by the Neo4j Graph Data Science library, enabling advanced network analyses.
TBEP Assistant
A distinguishing feature of TBEP is its integration with the TBEP Assistant, a specialized Question-Answering (QA) system. Built on a prompt fine-tuned large language model (LLM), the Assistant provides structured, citation-supported answers to biomedical questions. This enables researchers to extract mechanistic insights and generate hypotheses directly within the platform, bridging the gap between data analysis and biological interpretation.
For more information, please refer to our documentation.
